Mixed Finite Element Methods for Poisson's Equation
In this section, we consider solving the Poisson's equation using mixed finite element methods
\[-\nabla \cdot (\nabla u(x)) = f \text{ in } \Omega\tag{1}\]
with boundary conditions
\[u = g_D \text{ on } \partial \Omega\]
We reformulate the Eq. 1 as
\[\begin{aligned}\sigma + \nabla u &= 0 \\ \nabla \cdot \sigma & = f \end{aligned}\]
The mixed variational formulation is to find $(\sigma, u)\in H(\text{div}, \Omega) \times L^2(\Omega)$, such that
\[\begin{aligned}(\sigma, \tau) - (\nabla \cdot \tau, u) &= -(\tau\cdot \mathbf{n}, g_D)_{\partial \Omega}\\ (\nabla \cdot \sigma, v) &= (f, v)\end{aligned}\]
We use the following stable pair of elements: BDM${}_1$-P${}_0$, where the stress space is approximated using a BDM element while the displacement space is approximated using a piecewise constant element.
The code is as follows:
# Solves the Poisson equation using the Mixed finite element method
using Revise
using AdFem
using DelimitedFiles
using SparseArrays
using PyPlot
n = 50
mmesh = Mesh(n, n, 1/n, degree = BDM1)
testCase = [
((x, y)->begin
x * (1-x) * y * (1-y)
end,
(x, y)->begin
-2x*(1-x) -2y*(1-y)
end), # test case 1
((x, y)->begin
x^2 * (1-x) * y * (1-y)^2
end,
(x, y)->begin
2*x^2*y*(1 - x) + 2*x^2*(1 - x)*(2*y - 2) - 4*x*y*(1 - y)^2 + 2*y*(1 - x)*(1 - y)^2
end) ,# test case 2
(
(x, y)->x*y,
(x, y)->0.0
), # test case 3
(
(x, y)->x^2 * y^2 + 1/(1+x^2),
(x, y)->2*x^2 + 8*x^2/(x^2 + 1)^3 + 2*y^2 - 2/(x^2 + 1)^2
), # test case 4
]
for k = 1:4
@info "TestCase $k..."
ufunc, ffunc = testCase[k]
A = compute_fem_bdm_mass_matrix1(mmesh)
B = compute_fem_bdm_div_matrix1(mmesh)
C = [A -B'
-B spzeros(mmesh.nelem, mmesh.nelem)]
gD = bcedge(mmesh)
t1 = eval_f_on_boundary_edge(ufunc, gD, mmesh)
g = compute_fem_traction_term1(t1, gD, mmesh)
t2 = eval_f_on_gauss_pts(ffunc, mmesh)
f = compute_fvm_source_term(t2, mmesh)
rhs = [-g; f]
sol = C\rhs
u = sol[mmesh.ndof+1:end]
close("all")
figure(figsize=(15, 5))
subplot(131)
title("Reference")
xy = fvm_nodes(mmesh)
x, y = xy[:,1], xy[:,2]
uf = ufunc.(x, y)
visualize_scalar_on_fvm_points(uf, mmesh)
subplot(132)
title("Numerical")
visualize_scalar_on_fvm_points(u, mmesh)
subplot(133)
title("Absolute Error")
visualize_scalar_on_fvm_points( abs.(u - uf) , mmesh)
savefig("bdm$k.png")
endHere we have 4 test cases, and we show the results for each test case:
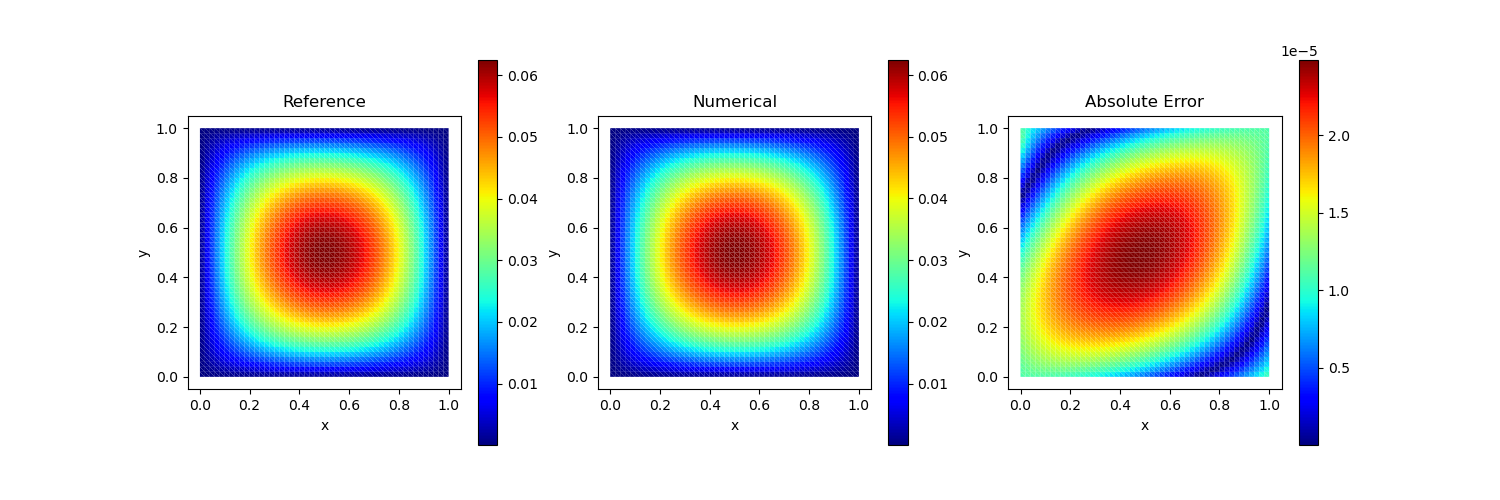
Test Case 1
Analytical solution:
\[u(x, y) = x(1-x)y(1-y)\]
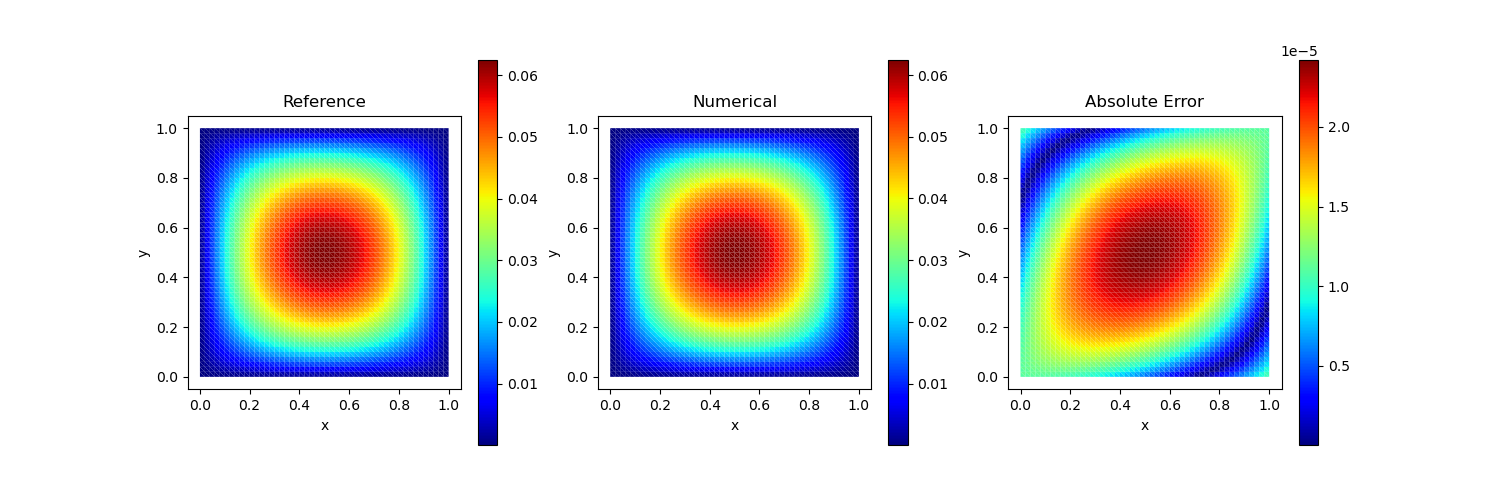
Test Case 2
Analytical solution:
\[u(x, y) = x^2 (1-x) y (1-y)^2\]
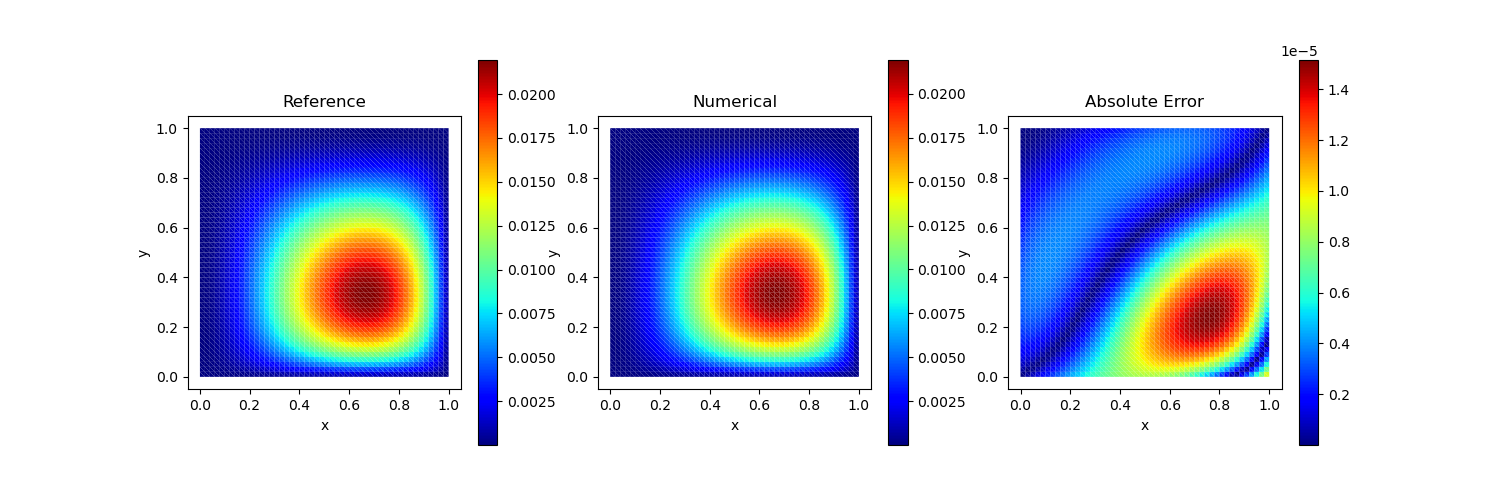
Test Case 3
Analytical solution:
\[u(x, y) = xy\]
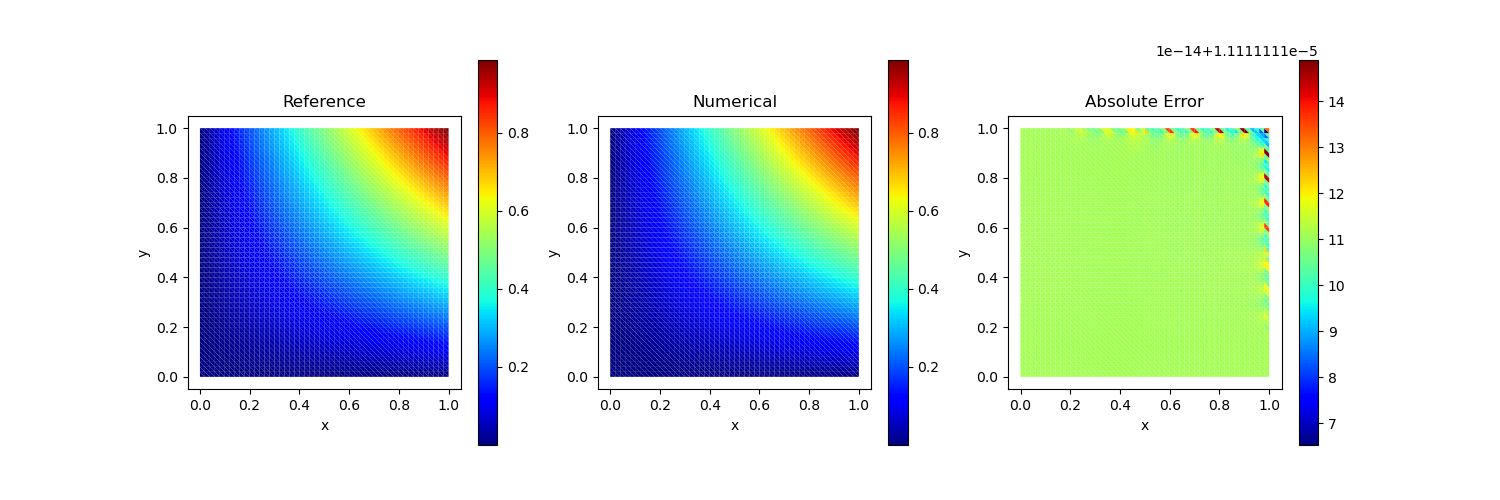
Test Case 4
\[u(x,y) = x^2 y^2 + \frac{1}{1+x^2}\]
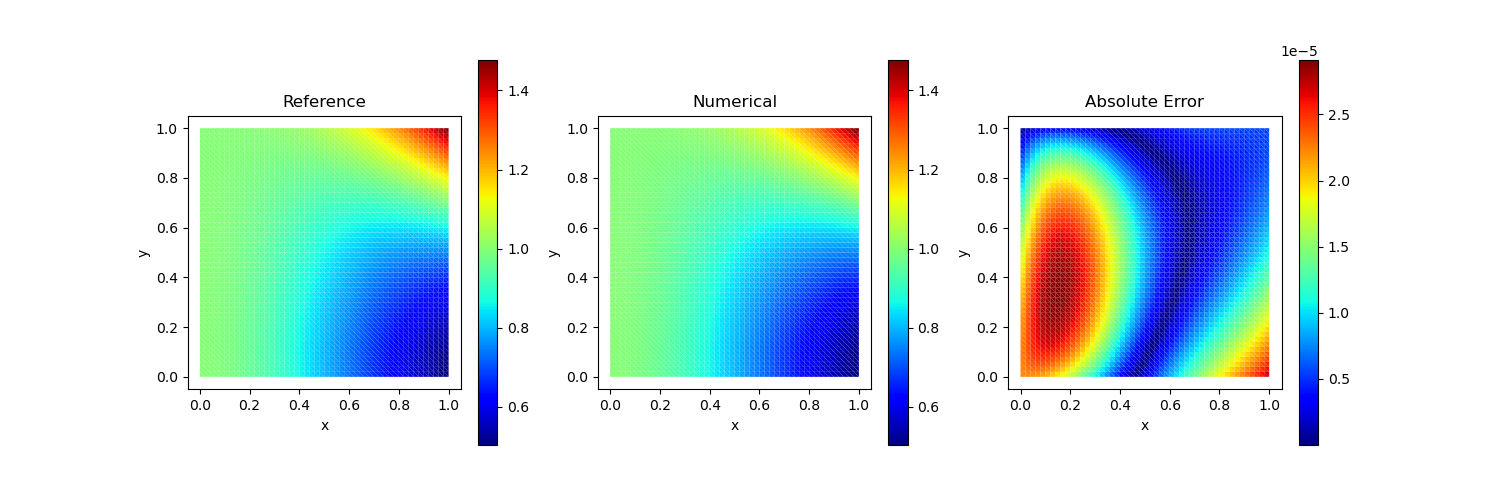
Traction Boundary Condition
We consider the case where the boundary conditions are given by
\[\sigma \cdot \mathbf{n} = g_N, x\in \Gamma_N, \quad u = g_D, x\in \Gamma_D\]
The weak form is
\[\begin{aligned}(\sigma, \tau) - (\nabla \cdot \tau, u) &= -(\tau\cdot \mathbf{n}, g_D)_{\Gamma_D}\\ (\nabla \cdot \sigma, v) &= (f, v)\end{aligned}\]
In this case, the traction boundary condition does not appear in the weak form, but determines the coefficients for the representation of $\sigma$ for the DOFs on the boundary $\Gamma_N$. Consider an edge $E_l = \{e_s, e_t\}, s<t$, then we have
\[\sigma = a\phi_{l,1} + b \phi_{l, 2}\]
which leads to
\[|E_l|g_N = a \lambda_s + b \lambda_t\]
To calculate $a, b$, we solve the following linear system
\[\begin{bmatrix}(\lambda_s, \lambda_s) & (\lambda_s, \lambda_t) \\ (\lambda_s, \lambda_t) & (\lambda_t, \lambda_t)\end{bmatrix}\begin{bmatrix}a\\b\end{bmatrix} = |E_l|\begin{bmatrix}(g_N, \lambda_s)\\ (g_N, \lambda_t)\end{bmatrix}\]
The boundary condition can be computed using impose_bdm_traction_boundary_condition1.
# Solves the Poisson equation using the Mixed finite element method
using Revise
using AdFem
using DelimitedFiles
using SparseArrays
using PyPlot
n = 50
mmesh = Mesh(n, n, 1/n, degree = BDM1)
function ufunc(x, y)
x * (1-x) * y * (1-y)
end
function ffunc(x, y)
-2x*(1-x) -2y*(1-y)
end
function gfunc(x, y)
(1-2x)*y*(1-y)
end
A = compute_fem_bdm_mass_matrix1(mmesh)
B = compute_fem_bdm_div_matrix1(mmesh)
C = [A -B'
-B spzeros(mmesh.nelem, mmesh.nelem)]
gD = (x1, y1, x2, y2)->!( (x1<1e-3 && y1>1e-3 && y1<1-1e-3) || (x2<1e-3 && y2>1e-3 && y2<1-1e-3))
gN = (x1, y1, x2, y2)->x1<1e-5 && x2<1e-5
D_bdedge = bcedge(gD, mmesh)
N_bdedge = bcedge(gN, mmesh)
t1 = eval_f_on_boundary_edge(ufunc, D_bdedge, mmesh)
g = compute_fem_traction_term1(t1, D_bdedge, mmesh)
t2 = eval_f_on_gauss_pts(ffunc, mmesh)
f = compute_fvm_source_term(t2, mmesh)
gN = eval_f_on_boundary_edge(gfunc, N_bdedge, mmesh)
dof, val = impose_bdm_traction_boundary_condition1(gN, N_bdedge, mmesh)
rhs = [-g; f]
C, rhs = impose_Dirichlet_boundary_conditions(C, rhs, dof, val)
sol = C\rhs
u = sol[mmesh.ndof+1:end]
close("all")
figure(figsize=(15, 5))
subplot(131)
title("Reference")
xy = fvm_nodes(mmesh)
x, y = xy[:,1], xy[:,2]
uf = ufunc.(x, y)
visualize_scalar_on_fvm_points(uf, mmesh)
subplot(132)
title("Numerical")
visualize_scalar_on_fvm_points(u, mmesh)
subplot(133)
title("Absolute Error")
visualize_scalar_on_fvm_points( abs.(u - uf) , mmesh)
savefig("mixed_poisson_neumann.png")The result is shown below